
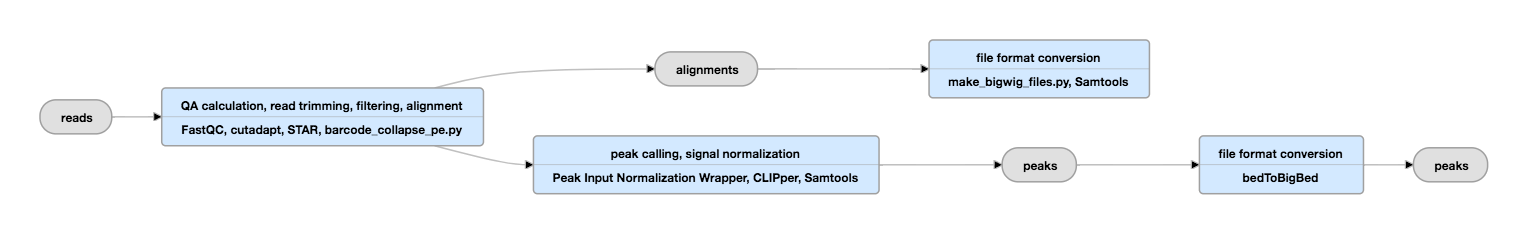

For more detailed information on NGS quality control, check out the tutorial for NGS here. Step 2: Quality ControlĪs for any NGS data analysis, CLIP-seq data must be quality controlled before being aligned to a reference genome. Open the Galaxy Upload Manager ( galaxy-upload on the top-right of the tool panel)įigure 1: Data can be imported directly with links.įigure 2: Imported datasets will appear in the history panel. Select the option Create New from the menu.Click on the galaxy-gear icon ( History options) on the top of the history panel.Tip: Creating a new historyĬlick the new-history icon at the top of the history panel. Step 1: Get data hands_on Hands-on: Data uploadĬreate and name a new history for this tutorial. We therefore want to process and validate the data from human liver cancer cells (HepG2) to find this conserved motif and in the process identify the function of RBFOX2 as well as describe the function of the targeted RNA. RBFOX2 is a relevant development and tissue-specific splicing factor with the conserved motif TGCATG. Inspection of peaks and aligned data with IGV.Generating bigWig files of the alignment ends.Functional analysis with RNA Centric Annotation System.Check the read coverage with plotFingerprint.Quality check of the de-duplication with FastQC.De-duplication with UMI-tools deduplicate.Step 4: Aligning Reads to a Reference Genome.Removal of adapter sequences with Cutadapt.Step 3: Removal of Adapters, Barcodes and Unique Molecular Identifiers (UMIs).Stranded means, if the read-library is strand specific, i.e., in a specific forward-reverse direction. Replicate is the number of the biological replicate. Hep G2 is a human liver cancer cell line. Table 1: Data from RBFOX2 CLIP-Seq and control experiment which we are going to use in this tutorial from a study published by Nostrand et al. Also included is a text file (.txt) encompassing the chromosome sizes of hg38 and a genome annotation (.gtf) file taken from Ensembl. The data was changed and downsampled to reduce data processing time, consequently the data does not correspond to the original source pulled from Nostrand et al. The dataset contains the first biological replicate of RBFOX2 CLIP-seq and the input control experiment (FASTQ files). The eCLIP data provided here is a subset of the eCLIP data of RBFOX2 from a study published by Nostrand et al.


 0 kommentar(er)
0 kommentar(er)
